4
HOW GENES FUNCTION
OBJECTIVES:
By the end of this session the
student should be able to:
* Use the genetic code to translate coding sequences
* Calculate the number of codons and amino acids from the number of
bases
* Name the main sites of non-coding DNA segments
* Name the function and features of the
promoter sequence
* Distinguish between introns and exons
* Explain how gene expression is controlled
THE GENETIC CODE
A gene is a stretch
of DNA that carries a coded message for the synthesis of a specific
protein. Most of the DNA of an organism
does not code for proteins. A stretch
of DNA is recognised as a gene coding for a protein if it is preceded by a promoter sequence. The promoter sequence is the site of
attachment for RNA polymerase, which is responsible for the process of
transcription of the genetic message to m-RNA. The promoter sequence is present
only on the template (transcribed) strand. The sequence of bases on m-RNA is
complementary to that on the transcribed strand of DNA but corresponds to that
on the coding strand except that T is replaced by U (Fig. 4.1).

The genetic code is, by convention, interpreted with reference to the sequence of bases on m-RNA. The m-RNA sequence happens to correspond to the coding strand of DNA, with the exception that U on RNA corresponds to T on DNA.
The sequence of
bases on m-RNA determines the exact sequence of amino acids in the protein. The
bases are read in triplets. Each triplet of bases is termed a codon and corresponds to a
particular amino acid. The genetic code designates how the codons correspond to
the amino acids. There are four base
(A, U, C and G) and 64 possible ways in which these can be combined to form codons
as shown in Fig. 4.2. Each codon is
specific for one amino acid. However,
there are only 20 amino acids and so one amino acid may be represented by more
than one codon. Because of this the genetic code is described as degenerate.
The codon AUG
codes for methionine but when it occurs after a promoter sequence it also
serves as a "start"
signal indicating the beginning of the coded message.
The codons UAA
(also called "ochre"), UAG (also called "amber") and UGA do not code for any amino acid but act as
"stop"
signals for the end of a gene message. Note that all the ‘stop’ codons start with U and include an A.

SIZES OF GENES AND
PROTEINS
Every three nucleotides correspond to one codon and one amino acid. If a
gene contains 1,200 nucleotides, this corresponds to 1,200/3 = 400 codons and
the resulting protein contains 400 amino acids.
The size of a protein is often expressed as its molecular mass. The molecular masses of amino acids vary but
it can be assumed that amino acids have an average molecular mass of
approximately 100 Daltons. Thus a
protein consisting of 400 amino acids will have a molecular mass of
approximately 40,000 Daltons. Conversely, a protein with a molecular mass of
60,000 Daltons is expected to contain about 60,000/100 = 600 amino acids and the gene coding for
this protein will consist of about 600 codons and 600 x 3 = 1,800 nucleotides.
THE C-VALUE PARADOX
The amount of DNA contained in a cell of a particular organism is termed
the C-value. Every organism has a
specific c-value. More complex
organisms are expected to contain more genes than simple organisms and consequently
would require more DNA. In fact,
however, the C-value or DNA content for a particular organism is not
proportional to the number of genes or to the complexity of the organism. Thus
the frog has seven times the DNA content of Man, and the Lilly has 100 times
the DNA content of Man. This is the C-value paradox.
It has been estimated that the human genome contains 3.5 billion base pairs. This amount of DNA could contain about 2 million genes. In fact, however, the total number of genes in man has been estimated to be only about 40,000 to 80,000. This means that about 3% of human genome codes for proteins, and 97 % of the human genome is non-coding DNA.
NON-CODING DNA
Although
over 95% of the total DNA is non-coding, it may have other important functions
such as the regulation of gene activity.
The main examples of non-coding DNA are the following:
1.
Promoter sequences
2.
Intervening
sequences (introns)
3.
Terminator sequences
4.
Sequences related to
chromosome structure
5.
Pseudogenes
6.
Repetitive DNA
These are explained in the following sections.
1. Promoter sequence
In order to be transcribed a gene must be preceded by a promoter sequence. This is
the recognition site for the attachment of RNA polymerase, the enzyme
responsible for transcription. The promoter sequence is followed by an initiator sequence,
which marks the site where transcription to m-RNA begins. The initiator codon or start
signal on m-RNA is the
sequence AUG, which is
also codon 1 and corresponds to the amino acid methionine. It also marks the site where translation
begins (Fig. 4.3).
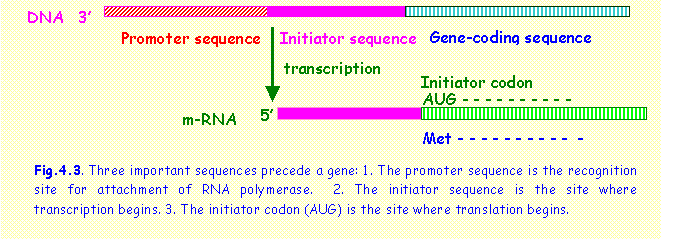
The initiator sequence is not the beginning
of the gene itself but the first part to be transcribed to m-RNA. The beginning of the gene itself is
indicated by the sequence "AUG" on m-RNA. This corresponds to “TAC” on the
transcribed strand. This is
codon 1 of the gene and is translated to the amino acid methionine. Subsequent triplets of bases are read as codons and are translated according to the genetic code until a “stop” signal is
encountered.
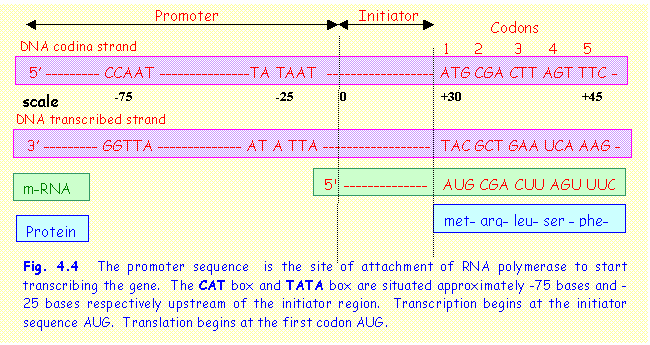
The promoter sequence
has two characteristic sequences by which it is recognised (Fig. 4.4). These
sequences are read as they occur on the coding
strand of DNA in the 5’ to 3’ direction.
1. The "TATA box" is a characteristic sequence
that is often TATAATA, or a closely similar sequence consisting of A and T
bases. It occurs about 25 bases before
the start (upstream) of the initiator region.
This type of sequence occurs quite regularly, with little variation in most
organisms and so is referred to as a highly conserved region or consensus sequence.
2. The "CAT box"
is a sequence consisting of CCAAT or
a similar sequence, and occurs about 75 bases upstream
of the initiator region. This is also a
conserved consensus sequence.
Note that DNA sequences are continuous and that there are no gaps demarcating promoter, initiator and codons. In figures 4.3 and 4.4, the boxes indicating the different sequences and the spaces between the codons have been used here only for the sake of clarity.
2. Intervening
sequences (introns)
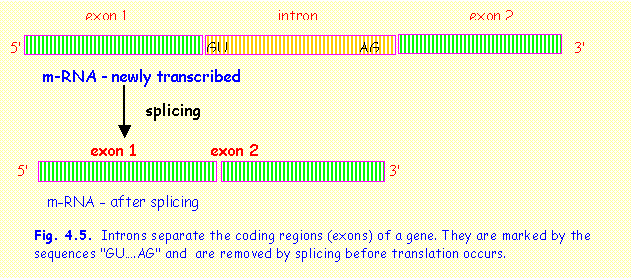
The
coded message of DNA is usually interrupted by one or more sequences that are
not part of the gene message. These intervening sequences are termed introns, while the actual coding sequences that are to be translated or
"expressed" are termed exons
(Fig.4.5). Introns are like blank spaces separating paragraphs in a written
message. They are transcribed on to the
newly formed m-RNA but are removed or "spliced"
before translation of the m-RNA begins. Splicing involves cutting out the
intron and re-fusion of the cut ends of the m-RNA. An intron usually begins with GU and ends with AG. These
sequences are recognition sites for splicing. The number of introns varies from 0 to 60 in
different genes. For a particular gene the number of introns is always the
same.
3. Terminator
sequences
A terminator sequence is one that causes RNA
polymerase to stop transcription. It occurs about 30 base pairs after the end
of the coding region of the gene. This
region usually contains the sequence AATAA or a closely similar sequence. Distinguish this from the terminator codon
(UAA, UAG or UGA) that marks the end of translation or the end of polypeptide synthesis.
4. Sequences related to chromosome structure
These are other
non-coding sequences that have the following functions:
a) Sequences
marking the telomeres (chromosome ends);
b) Sequences
marking the centromeres and the kinetochores for attachment of the mitotic spindle;
c) Sequences
marking sites where DNA replication begins;
many of these sites occur at intervals along each chromosome.
5. Pseudogenes
These
are sequences that are similar to the sequences of normal genes, but lack a
promoter region and therefore are non-functional or“false” genes. They are the
result of evolution. Genes, which had been at one time functional, had lost their
promoter sequence through mutations during the process of evolution, but have
remained in the genome as "pseudogenes". Usually, they are situated close to normal genes having a similar
sequence. Examples are seen in the
globin genes of hemoglobin.
6. Repetitive DNA
These are sequences
that are repeated many times over. Some of these are trinucleotide repeats e.g. CAG CAG CAG … etc
repeated many times over. They are often present in non-coding regions of the
genome, but sometimes they are part of a coding region of a gene and are
translated into amino-acids e.g. CAG repeats in the example above translate to “polyglutamine”
repeats in the protein.
Other examples are di-nucleotide repeats,
which are sometimes repeated hundreds of times. Some telomere and centromere
sequences are repetitive DNA sequences.
CONTROL OF GENE
EXPRESSION
Although all cells of
an individual contain the same genes they are not all functional at the same
time. The genes that are active in a
particular cell depend on the cell type and its state of activity. The genes that are active in brain cells are
different from those that are active in liver, kidney, muscle, fibroblasts
etc. Furthermore, some genes are active
only during embryonic development and are switched off in the mature
tissues. How are genes switched on an
off?
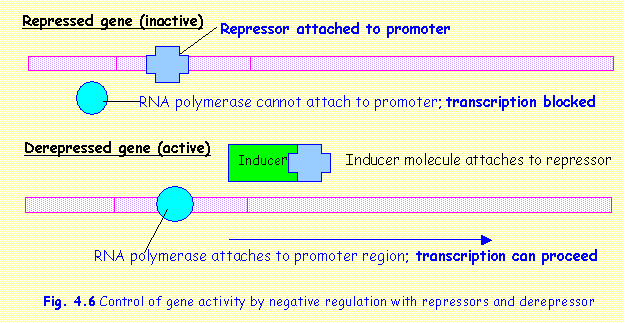
In
any one particular cell most genes are inactive and only those relevant to the functioning
of the particular cell are active. As noted above, the promoter region binds to
RNA polymerase, which then transcribes m-RNA.
In inactive genes the binding of RNA polymerase is prevented by means of
a repressor, a
molecule that attaches itself to the promoter region, blocking the RNA
polymerase binding site (Fig.4.6).
Activation of the gene requires removal of the repressor. This is
achieved by an inducer,
which combines with the repressor and prevents it from binding to DNA. This is termed negative regulation of gene activity. Most genes are normally inactive, or repressed, and are switched
on or de-repressed, only when required.
Activators may also control gene activity. These molecules enhance the binding of RNA
polymerase to the promoter region and therefore exert positive
control on
transcription.
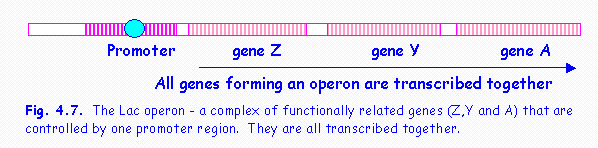
OPERONS
In some cases a
group of functionally related genes are situated next to one another on the
same DNA strand, and share the same promoter region. Such a unit is termed an operon. An example is the lac operon in bacteria. This operon
contains three genes (called Z,Y and A) that are responsible for three enzymes
involved in different stages of lactose metabolism. The three genes are situated next to one another, are transcribed
together and are controlled by one promoter. When the promoter region is
repressed all three genes are switched off.
PROBLEMS
1. The following sequence is part of the
transcribed strand of DNA:
3' G C
T A A T C A
G T
G C G T A 5'
Write the sequences of:
a.
the coding strand of
DNA
b.
m-RNA
c.
amino acids
2. The following is a schematic diagram of DNA being transcribed:
a.
 DNA transcribed strand
DNA transcribed strand
b.
DNA
coding strand
c.
m-RNA
The direction of transcription is shown.
Label the 5' and 3' ends of each strand.
3. The following is the sequence
of amino acids of the last part of a protein molecule.
Using the genetic code, write the codons on m-RNA and label the 3' and 5' ends.
(Where more than one codon is possible use the first one in the table.) Then, write the corresponding codons on the
template strand of DNA.
Protein
m-RNA - arg - leu - asp - phe - pro - ile - glu - gly - val -DNA
template
strand