The Nucleolus, Ribosomes and Protein Synthesis
Professor
Alfred Cuschieri
Department
of Anatomy, University of Malta
Objectives
![]() State the main
characteristics of messenger, ribosomal and transfer RNA
State the main
characteristics of messenger, ribosomal and transfer RNA
![]() Distinguish between
transcription and translation
Distinguish between
transcription and translation
![]() Describe the composition and
structure of the nucleolus
Describe the composition and
structure of the nucleolus
![]() Explain what constitutes the
nucleolus organizer regions
Explain what constitutes the
nucleolus organizer regions
![]() Outline the role of ribosomes
in protein synthesis
Outline the role of ribosomes
in protein synthesis
![]() Discuss how antibiotics
inhibit the growth of bacteria and have different sites of action
Discuss how antibiotics
inhibit the growth of bacteria and have different sites of action
Recommended reading
The
World of the Cell: Becker WM, Kleinsmith LJ, Hardin J. 4th Edition
![]() Chapter 19 Gene Expression:
I. The Genetic Code and Transcription
Chapter 19 Gene Expression:
I. The Genetic Code and Transcription
![]() Chapter 20 Gene Expression:
II. Protein Synthesis and Sorting
Chapter 20 Gene Expression:
II. Protein Synthesis and Sorting
The nucleolus and ribosomes form part of the protein synthesizing machinery of the cell
![]() The nucleolus is the site where most of the ribosomal RNA (r-RNA)
is transcribed
The nucleolus is the site where most of the ribosomal RNA (r-RNA)
is transcribed
![]() Ribosomes are composed of r-RNA and proteins
Ribosomes are composed of r-RNA and proteins
![]() The ribosomes are the sites where protein synthesis occurs
The ribosomes are the sites where protein synthesis occurs
![]() Synthesis of specific proteins (transcription) also requires the action
of two other types of RNA:
Synthesis of specific proteins (transcription) also requires the action
of two other types of RNA:
![]() m-RNA as a
template
m-RNA as a
template
![]() t-RNA for the
assembly process
t-RNA for the
assembly process
The Cell Has 3 Types of RNA
![]() messenger
RNA (m-RNA)
messenger
RNA (m-RNA)
-
there are about 105 varieties; each corresponds to a gene; each carries a coded message to cytoplasm
![]() ribosomal
r-RNA (r-RNA)
ribosomal
r-RNA (r-RNA)
there are 4 varieties; 4 RNA constituents of ribosomes
![]() transfer
t-RNA (t-RNA)
transfer
t-RNA (t-RNA)
there are 20 varieties corresponding to 20 amino-acids; they transfers amino acids to polypeptides chain
m-RNA is the transcript
of a gene
DNA has two strands. The template strand carries the message and is transcribed to m-RNA.
5' --- T G T A C G A T T C C G A T G A C T --------3' coding strand
3' --- A C A T G C T A A G G C T A C T G A --------5' template strand
![]() 5' --- U G U A C G A U U C C G A U G A C U --------5' m-RNA
5' --- U G U A C G A U U C C G A U G A C U --------5' m-RNA
codon codon codon codon codon codon
A1 --- A2 --- A3--- A4 --- A5 --- A6----- amino
acid chain (Polypeptide)
Note:
![]() M-RNA is translated to
protein. A triplet of bases on m-RNA is a codon and corresponds to an amino
acid.
M-RNA is translated to
protein. A triplet of bases on m-RNA is a codon and corresponds to an amino
acid.
![]() m-RNA is similar to the coding strand except that T is replaced by
U.
m-RNA is similar to the coding strand except that T is replaced by
U.
![]() transcription occurs in the 5' to 3' direction; nucleotides are always added at the 3 end
transcription occurs in the 5' to 3' direction; nucleotides are always added at the 3 end
![]() translation also proceeds in the 5to 3
direction
translation also proceeds in the 5to 3
direction
Transcription
![]() Is the process whereby the genetic message for
a specific protein is transcribed on to m-RNA using the transcribed strand of
DNA as template.
Is the process whereby the genetic message for
a specific protein is transcribed on to m-RNA using the transcribed strand of
DNA as template.
![]() Occurs
in the nucleus of eukaryotic cells
Occurs
in the nucleus of eukaryotic cells
![]() Occurs in the 5 to 3 direction
Occurs in the 5 to 3 direction
![]() The sequence of bases on m-RNA:
The sequence of bases on m-RNA:
![]() Is complementary to that on the transcribed
strand of DNA
Is complementary to that on the transcribed
strand of DNA
![]() Corresponds
to that on the coding strand except that U replaces T
Corresponds
to that on the coding strand except that U replaces T
Translation
![]() Is the process of synthesis of a specific protein using m-RNA as a
template.
Is the process of synthesis of a specific protein using m-RNA as a
template.
![]() Occurs in the ribosomes in the cytoplasm
Occurs in the ribosomes in the cytoplasm
![]() The message is read in codons (triplets of
nucleotides) in the 5 to 3 direction
The message is read in codons (triplets of
nucleotides) in the 5 to 3 direction
![]() Each codon corresponds to an amino acid
according to the genetic code
Each codon corresponds to an amino acid
according to the genetic code
THE
GENETIC CODE
![]() The genetic code is, by convention, interpreted with
reference to the sequence of bases on m-RNA.
The genetic code is, by convention, interpreted with
reference to the sequence of bases on m-RNA.
![]() The sequence of bases on m-RNA corresponds to that on
the coding strand of DNA, except that U in RNA replaces T on DNA.
The sequence of bases on m-RNA corresponds to that on
the coding strand of DNA, except that U in RNA replaces T on DNA.
![]() The sequence of bases on m-RNA determines the exact
sequence of amino acids in the protein.
The sequence of bases on m-RNA determines the exact
sequence of amino acids in the protein.
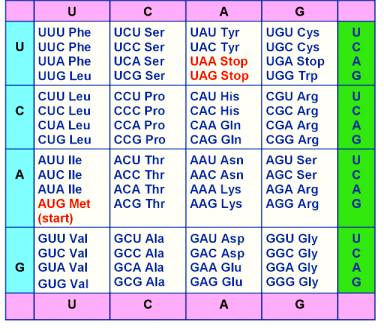 The Genetic
Code
The Genetic
Code
![]() Note that there are:
Note that there are:
4 bases - A, U, C, G;
64 possible codons;
20 amino acids
![]() The genetic code is degenerate i.e. One amino acid may be
represented by more than one codon
The genetic code is degenerate i.e. One amino acid may be
represented by more than one codon
![]() The codon AUG codes for methionine but it may also serve
as a "start" signal indicating the beginning of the
coded message.
The codon AUG codes for methionine but it may also serve
as a "start" signal indicating the beginning of the
coded message.
![]() The codons UAA,
UAG, UGA do not
code for any amino acid but act as "stop" signals for the end of a gene message.
The codons UAA,
UAG, UGA do not
code for any amino acid but act as "stop" signals for the end of a gene message.
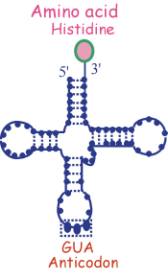
t-RNA
![]() Consists of a single strand of RNA folded in the form of a cross
Consists of a single strand of RNA folded in the form of a cross
![]() Has an anti-codon at one end (triplet complementary to sequence of
a codon)
Has an anti-codon at one end (triplet complementary to sequence of
a codon)
![]() Has the corresponding amino acid at the opposite end
Has the corresponding amino acid at the opposite end
In this example the anticodon GUA corresponds to the amino acid histidine and the codon CAU on m-RNA
r-RNA
![]() Is involved in the bio-synthesis of ribosomes together
with proteins
Is involved in the bio-synthesis of ribosomes together
with proteins
![]() 4 types of r-RNA: 5S, 5.8S, 18S, 28S (distinguished by the sedimentation coefficient)
4 types of r-RNA: 5S, 5.8S, 18S, 28S (distinguished by the sedimentation coefficient)
![]() is transcribed from multiple copies of DNA
is transcribed from multiple copies of DNA
(unlike m-RNA transcribed from a unique
gene)
![]() is synthesised in the nucleolus
is synthesised in the nucleolus
The Nucleolus
Consists
of two parts:
-
fibrillar
part - consists of chromatin:
(DNA transcribing r-RNA)
-
granular
part consists of ribonucleoprotein particles
(r-RNA + proteins)
The nucleolar DNA
![]() The genes for 5.8S, 18S and 28S r-RNA
form a gene cluster
The genes for 5.8S, 18S and 28S r-RNA
form a gene cluster
![]() they are all transcribed together
forming a r-RNA complex of 45S
they are all transcribed together
forming a r-RNA complex of 45S
![]() these genes are present on the nucleolus organiser regions on the
satellites of the acrocentric chromosomes 13, 14, 15, 21 and 22
these genes are present on the nucleolus organiser regions on the
satellites of the acrocentric chromosomes 13, 14, 15, 21 and 22
![]()
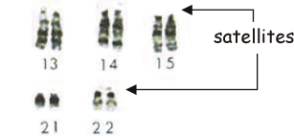
Note: The gene
for 5S r-RNA is located on chromosome 1 and is transcribed separately
The nucleolus organiser regions contain
many repeated copies of the r-RNA gene cluster
![]()
![]() This
is an example of gene amplification
This
is an example of gene amplification
![]() Multiple
copies of r-RNA are transcribed simultaneously
Multiple
copies of r-RNA are transcribed simultaneously
![]() This
forms a feather arrangement: the stem
is the transcribed strand of DNA; the side strands are the forming RNA of various lengths depending on how much of
the strand has been transcribed.
This
forms a feather arrangement: the stem
is the transcribed strand of DNA; the side strands are the forming RNA of various lengths depending on how much of
the strand has been transcribed.
Procedure for Isolation of Ribosomes
1.
Prepare cell homogenate
2. Differential
centrifugation to separate RER layer
3. Treat with detergent to remove membranes
4. Differential
centrifugation to separate ribosomes
from debris
5. Treat with low [Mg2+] - cleaves ribosomes into small and large
sub-units
Composition
of Ribosomes
80 S ribosomes can be broken down into
two sub-units by adjusting Mg 2+ concentration:
 Small sub-unit
- 40 S
Small sub-unit
- 40 S
Large sub-unit - 60 S
Structure
of a Ribosome
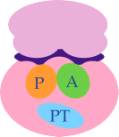
Groove for binding of m-RNA
Amino-acyl site (A) for binding to next
t-RNA
Peptidyl transferase site (PT)
for binding of amino acids by peptide bonds
Peptidyl site (P) for the growing
peptide chain
Protein
synthesis involves a number of steps:
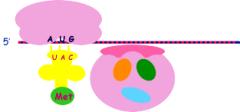
1. Initiation
a.
Dissociation of ribosome subunit requiring an initiation factor and energy
derived from  GTP
GTP
b. Formation
of an initiation complex consisting of the small subunit and the first t-RNA.
The initiator
codon is AUG
and the initial
amino acid is methionine
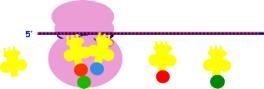
c. The
ribosome is closed
-
the
initiation complex has a one amino acid
(met) attached to it
2. Translocation
m-RNA moves by one codon; a new t- RNA occupies the A site;
the two amino acids fuse at the PT site
3. Chain elongation
m-RNA moves by one codon; t- RNA is displaced from the P site; another enters
the A site
Elongation of
the polypeptide chain requires an elongation factor and GTP
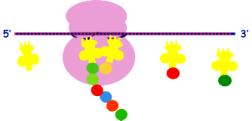
4. Chain termination
A releasing
factor is required for ending protein synthesis. It attaches to the terminator codon UAG.
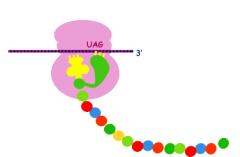
Polyribosomes
 Free ribosomes usually occur in small
clusters termed polyribosomes.
Free ribosomes usually occur in small
clusters termed polyribosomes.
UsuallyOneOne m-RNA runs successively through the several ribosomes in a cluster
producing multiple copies of a protein
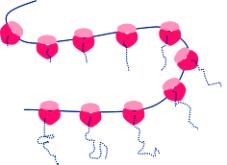
Ribosomes may attach to
the RER
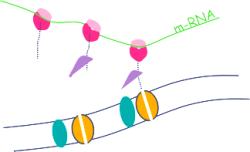 Attacment of ribosomes to RER requires:
Attacment of ribosomes to RER requires:
1. A signal peptide the first part of the protein being transcribed
2. A signal recognition particle (SRP) attaches to the signal protein
3. A docking protein to bind to the SRP
4. Ribophorin - a membrane protein forms a channel for the
signal peptide to enter the lumen of the
RER
Many
Antibiotics act by Inhibiting Protein Synthesis in Bacteria.
Bacterial (prokaryotic) ribosomes are similar in their
action to mammalian ribosomes but differ their sedimentation coefficient (70S consisting
of 30S and 50S subunits), the details of the r-RNA and of the associated
proteins .
Antibiotics
act at various sites in the protein synthesis pathway. The following are some examples:
![]() Inhibiting the attachment of the first t-RNA to the small subunit
Inhibiting the attachment of the first t-RNA to the small subunit
e.g.Streptomycin
![]() Iinhibiting the binding of t-RNA to A-site
Iinhibiting the binding of t-RNA to A-site
e.g.Tetracycline; Fusidic
acid
![]() Blocking
protein synthesis by bind to the A site
(molecular analogue of t-RNA)
Blocking
protein synthesis by bind to the A site
(molecular analogue of t-RNA)
e.g. Puromycin
![]() Preventing
ribosome assembly by binding to the large (50S) subunit
Preventing
ribosome assembly by binding to the large (50S) subunit
e.g.
Eryhthromycin
![]() Inhibition
of peptidyl transferase site
Inhibition
of peptidyl transferase site
e.g. Chloramphenicol ; Lincomycin
![]() Prevents translocation of the protein from A to P site
Prevents translocation of the protein from A to P site
e.g.Thioseptrin
Why do antibiotics inhibit protein synthesis in bacteria but not in humans?
![]() Many antibiotics are specific for prokaryotic
(bacterial) ribosomes (70S)
Many antibiotics are specific for prokaryotic
(bacterial) ribosomes (70S)
![]() Prokaryotic ribosomes (70S) have a
different r-RNA and protein composition from eukaryotic (80S) ribosomes
Prokaryotic ribosomes (70S) have a
different r-RNA and protein composition from eukaryotic (80S) ribosomes
![]() The
bacterial cell wall is permeable to some antibiotics while the plasma membrane
of eukaryotic cells is not
The
bacterial cell wall is permeable to some antibiotics while the plasma membrane
of eukaryotic cells is not
Some inhibitors of protein synthesis also act on eukaryotic cells
![]() Substances
that inhibit protein synthesis in human cells are lethal to cells that
are actively dividing
Substances
that inhibit protein synthesis in human cells are lethal to cells that
are actively dividing
![]() Such
substances are useful as cytostatic
agents e.g. cycloheximide
Such
substances are useful as cytostatic
agents e.g. cycloheximide
*************