
Free Radicals
A fraction of the molecular oxygen generated in respiring cells is subjected
to sequential univalent reduction to superoxide radicals (O2
-), hydrogen peroxide (H2O2), and hydroxyl radicals.Unless
effectively removed, these highly reactive oxygenspecies cause damage to DNA
, proteins, and lipids.To protect their cellular components, organisms have
evolved a series of chemical and enzymatic mechanisms that removethese detrimental
oxygen species.
In the first line of defense is the family of superoxide dismutases (SOD,1
EC. 1.15.1.1),which scavenge superoxide anions and catalyze their dismutationinto
hydrogen peroxide and oxygen :
2O2- + 2H + ------> H2O2 + O2
Free radicals have been directly and indirectly implicated inthe aging process. A transgenic strain of Drosophila mela-nogasteroverexpressing both SOD and catalase exhibited alife-extension of up to one-third that of wild-type. Long-lived genetic variants of the metazoan Caenorhabditis elegans,age-1 and daf-2, provide evidence for a positive correlation between cellular levels of SOD and extended lifespan.
Superoxide Dismutase
Four genetically distinct forms of SOD occur that differ in their active site
prosthetic metal ion and/or cellular localizalion.
The metals these metalloenzymes utilise in their active sites are iron (FeSOD),
manganese (MnSOD), nickel (NiSOD) or copper and zinc (CuZnSOD). The nickel-containing
enzyme is rare, and has been shown to be present in only one organism.The
metal is their to undergo a cyclic change in oxidation state during the catalytic
dismutation reaction. In CuZnSOD it is the Cu which undergoes this cyclic
process:
M3+ + O2- ------> M2+ + O2 i
M2+ + O2 - + 2H+ ------> M3+ + H2O 2 ii
where M is the metal, Fe, Mn, Ni or Cu
Eukaryotic cells possess a cytosolic CuZnSOD, a glycosylated extracellular
CuZnSOD, and a mitochondrial MnSOD.
Most prokaryotes generally have both MnSOD and FeSOD that are similar in
their amino acid sequence and structure. Many prokaryotes also posess a CuZnSOD
which is found in the periplasmic space.
Mitochondrial MnSOD is encoded by a nuclear gene and isproduced as a precursor
protein targeted to the mitochondrialmatrix during which it is processed
into the mature form by cleavage of the transit peptide. The expression of
MnSODis usually inducible in both prokaryotic and eukaryotic cells, its levels
increasing under conditions of elevated oxidative stress in the immediate
environment .
RESEARCH:
We are investigating the differences between gene expression
in the nematode C.elegans under a variety of conditions which produce
oxidative stress in this multicelled, eukaryotic organism.
Structure
Information about the three dimensional structure of protein molecules is
readily available and the number of structures solved as evidenced by the
number of entries in the Brookhaven database is expanding at an exponential
rate.
Knowing the structure of a
particular protein is of tremendous use when designing experiments to better
understand its biological function or catalytic mechanism at a sub-molecular
level. A minimum requirement to beginning structure-function studies is to
determine the primary structure (sequence) of one's protein. If no X-ray
or NMR structural data are available structures of proteins with homologous
sequences are often adequate.
MnSOD-3 from Caenorhaditis elegans
We have previously isolated and sequenced the gene and mRNA for both SOD-2 and ASOD-3 of the nematode worm, C. elegans. In a collaboration with the University of Leeds, UK we have now solved their structures too. Below is a representation of the tetrameric MnSOD SOD-3.
The x-ray diffraction and data collection was performed at the new Diamond Light Source ( click here for an image)

human CuZnSOD
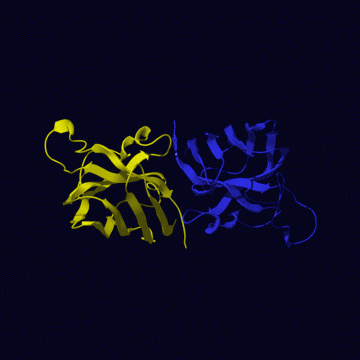
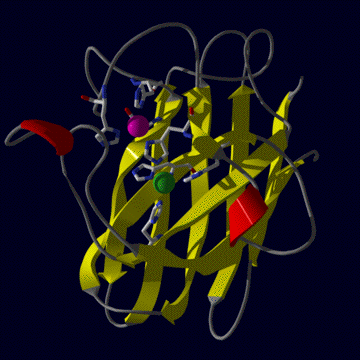
The human CuZnSOD exists in a dimeric, cytosolic form
(left figure) and a tetrameric extracellular form. The tertiary structure
(right figure) is mainly beta sheet (shown in yellow), only one small helix
is present (two helical regions are shown in red). Only dimers are active,
although the equivalent homologous enzyme from E.coli periplasm is
a monomeric enzyme. The two metals (copper is green zinc is purple) are locked
in place by seven liganding residues (one aspartic acid and six histidines)
but each metal is coordinated by four. Thus one residue, a histidine, bridges
between the two metal ions.
RESEARCH:
We are investigating the differences between human
and E.coli CuZnSODs. By changing the protein structure of human CuZnSOD and
targetting the expressed protein in the periplasmic space of E.coli
cells, we aim to see if a monomeric SOD can be produced from the human enzyme.
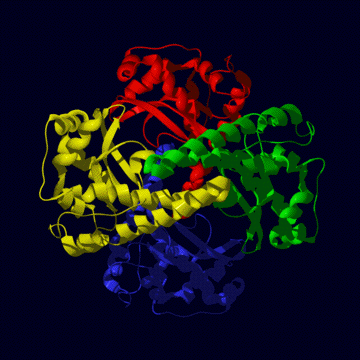
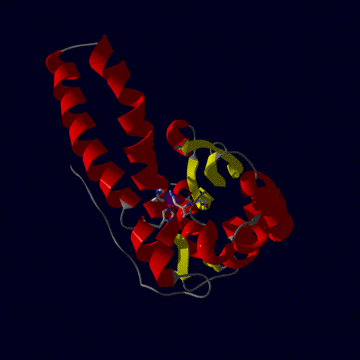
human MnSOD
The human mitochondrial enzyme MnSOD exists as a tetrameric
protein (left figure) , each subunit of which binds one atom of manganese
in the fully-metallated state. To be active, the metal must be in the Mn
III state (see reactin i above). The tertiary structure (right figure) is
one of alpha plus beta, mostly alpha helix (red) but with three beta strands
making a beta sheet (yellow). The metal (purple sphere) is held in position
by three histidine and one aspartic acid residue.
RESEARCH:
The effect of chaperone proteins on the imported precursor
form of MnSOD is being compared with that of the mature form of the enzyme.
We have already discovered different effects of chaperonins on the E.coli
enzymes.
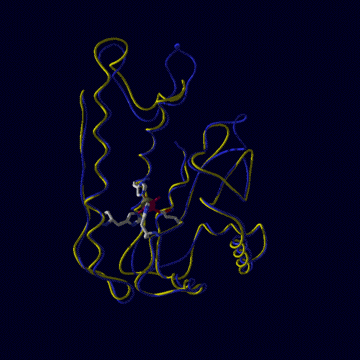
E.coli MnSOD and FeSOD
E.coli contains both MnSOD and FeSOD. As well as having a high degree of
homology between themselves (in the left figure, FeSOD is yellow, MnSOD is
blue), all mononuclear SODs like these are all very similar on a stuctural
level, though some are active as dimers (as in E.coli) while others
are tetramers (as in human, above). For example compare the folded shape of
human MnSOD above with the FeSOD and MnSOD of E.coli (below left).
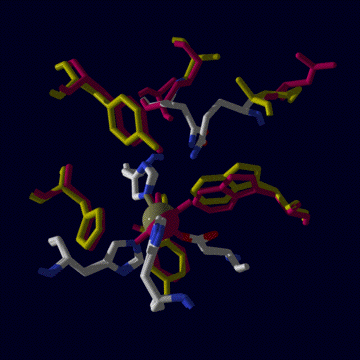
Given this high degree of similarity, it has been difficult by examination
of phylogenetic data using protein sequence information and structural data
to ascertain the nature of metal specificity in these enzymes. Why does E.coli
MnSOD only fold around manganese and FeSOD only around iron? Why are these
active only when the correct metal is present in their active site? Active
sites are compared in the right figure, below. The only major difference
between them is the glutamine residue at the top of the figure. In FeSOD
it derives from the N domain, in MnSOD it comes from thye C domain.
RESEARCH:
We are investigating the nature of metal selectivity
and specificity in these enzymes. Using site-directed mutagenesis we have
produced an FeSOD with glutamine shown above in the characteristic orientation
of MnSOD. We have also carried out the same reactions with MnSOD so that the
glutamine residue resembles the FeSOD.